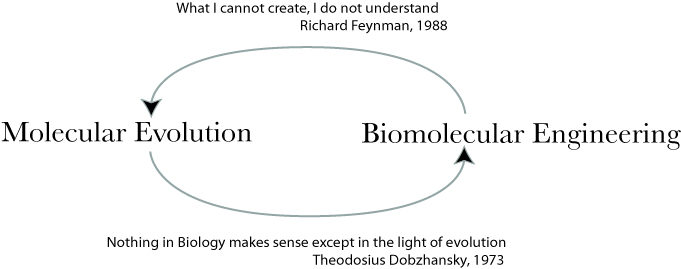
Directed Evolution - REAP (click for schematic)
Directed evolution and protein engineering often utilize sequence information contained within homologs to guide the design of diverse libraries. Combinatorially capturing this information can require the production of 106-1012 variants in a library. The ability to cull the sequence information thus holds considerable value. We have developed a new evolutionary-guided approach for directed evolution and protein engineering, Reconstructing Evolutionary Adaptive Paths (REAP). The approach culls sequence information based on phylogenetic analysis of homologous sequences, detection of functional divergence among lineages of the phylogeny and reconstruction of the individual mutations that occur along branches of the phylogeny inferred to have undergone functional divergence. REAP analysis highlights sites implicated in functional divergence or adaptive evolution during the history of a gene family. This provides an opportunity to disregard the vast number of neutrally evolving sites contained within homologous sequences when designing libraries. The utility of the REAP approach was recently demonstrated by engineering DNA polymerases to faithfully incorporate a modified nucleoside triphosphate useful for next-generation DNA sequencing technology.
Reconstruction and resurrection of ancient sequences
Ancestral sequence reconstruction (ASR) consists of inferring ancestral character states of DNA and protein sequences when analyzed within a phylogenetic context. Proposed by Linus Pauling in 1963 and first put into practice separately by Steve Benner and Allan Wilson in 1990, ASR provides a unique way to travel through time to understand paleoenvironments of ancient organisms, functional diversification of biomolecules and regulatory pathways. Our laboratory applies ASR to gene families such elongation factors, thioredoxins, ribosomal proteins, uricases, inter alia.
Experimental evolution
Experimental evolution consists of evolving microorganisms in the laboratory in attempt to understand how genotype and phenotype are linked within functional/adaptive landscapes. We are using our resurrected elongation factor proteins inserted into E. coli as a system to study evolutionary trajectories in landscapes. In particular, we are testing whether replaying the 'tape of life' is repetitive and whether historical contingency funnels molecular trajectories.
Experimental phylogenetics
Experimental phylogenies consist of evolving microorganisms in the laboratory in a manner consistent with the phylogenetic bifurcation that describes how natural organismal evolve. This technique allows us to generate a 'known' phylogeney in the laboratory in which we determine the properties of the phylogeny (such as branching patterns, branch lengths, functional diversification, etc.). We are using the fluorescent protein gene family to generate known phylogenies, with known ancestral states, in our attempt to benchmark ancestral sequence reconstruction.
Engineering proteins composed of unnatural amino acids
Our laboratory exploits models of molecular sequence evolution to guide us in our attempt to engineer novel biomolecular systems. For instance, our lab is engineering tRNAs and Elongation Factors biomolecules to participate in a synthetic, tunable protein translation system. The goal here is to generate peptides and proteins that are composed of amino acids different than the 20 canonical amino acids. This is a particularly exciting synthetic system to develop based on the biomolecular emergent properties we expect to generate.
Synthetic/Ancestral genomes
We anticipate that the synthesis of recombinant, minimal and/or ancestral genomes will have a profound effect on our understanding of early life. As molecular reconstructionists, we are interested in constructing a complete ancestral biochemical pathway (e.g., operon), or even a complete ancestral genome? The ancestral reconstruction field would no longer be confined to single gene analysis. It is also quite possible that our understanding of what constitutes a sustaining minimal genome required to support life will be altered through ancestral reconstructions. In this way, homologous genes performing two different, but related, functions may share a single common ancestor that performed both of these functions, albeit with less efficiency or specificity.
Models of molecular sequence evolution
Our laboratory has developed models to better understand functional divergence among homologous sequences. One such model is a modification of Walter Fitch's covarion model first described in the 1970's. We have shown how this model performs under phylogenetic conditions such as parsimony and maximum likelihood. Much of our work has involved analyzing the Elongation Factor-Tu gene family using computational analysis. We are currently attempting to experimentally validate the computational predictions of our models.
Evolutionary Biomedicine
There is a recent appreciation that biomedicine can be better served by incorporating evolutionary analysis into the field. This appreciation stems from the realization that evolutionary or historical explanations of the DNA and proteins responsible for diseased states provide insight into susceptibility of mutations. Our laboratory attempts to extend the application of evolutionary models into the biomedical field. We combine models of molecular sequence evolution and ancestral sequence reconstruction to engineer biomolecules for therapeutic purposes.
Origins of the ribosome and the genetic code
The goal here consists of computationally reconstructing components of an ancient ribosome, resurrecting them in the laboratory, and assaying for their ability to participate in protein translation (peptidyltransferase activity) in combination with modern and ancient (or minimal) ribosomal RNA structures. We anticipate that this approach will enable us to computationally dissect and experimentally reassemble the ribosome in a manner that allows to understand the origins and early evolution of peptide synthesis.
Evolution and engineering of protein thermostability
Protein engineering and directed evolution are powerful techniques for improving the activity, specificity and/or stability of proteins. Enhancing the stability of proteins is one of the most common goals in directed evolution studies. The approach we are developing attempts to incorporate evolutionary information similar to the other approaches, but our approach uniquely incorporates this information within the context of a historical model (phylogenetic tree). This will enable us to follow evolutionary trajectories of adaptation (in particular, thermostability), dissect additive versus epistatic biophysical interactions and delineate neutral versus adaptive changes - all within a framework of protein structure.






