- Home
Welcome !
Welcome to john houghton's home page for his biology courses. This site is designed as a hub for curating and sharing lectures, course syllabi, assignments, and links to relevant resources. Use the menu bar at the top of the screen to navigate through the site.
(Please note: this page is currently under construction.)

- BIOL 2107
Fall '22 CRN87989
Lectures: (1)
- Courses
- Resources
General Resources

DNA: base_pairing -Animation RNA Transcription I -Animation
DNA Replication -Animation RNA Transcription II -Animation
The CENTRAL DOGMA..... revisited
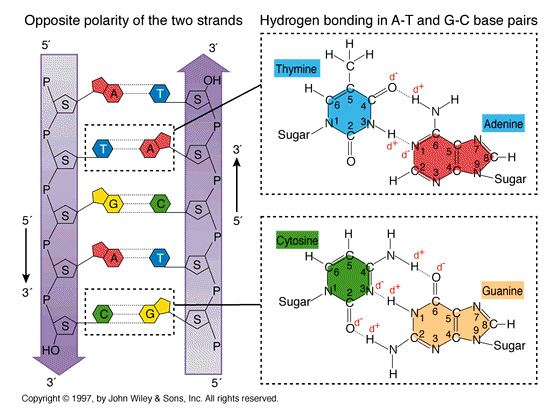
The DNA molecule is a double-stranded helix.The diameter of the DNA molecule is uniform.
The twist in DNA is right-handed (the twist is in the same direction as the thread on most screws).
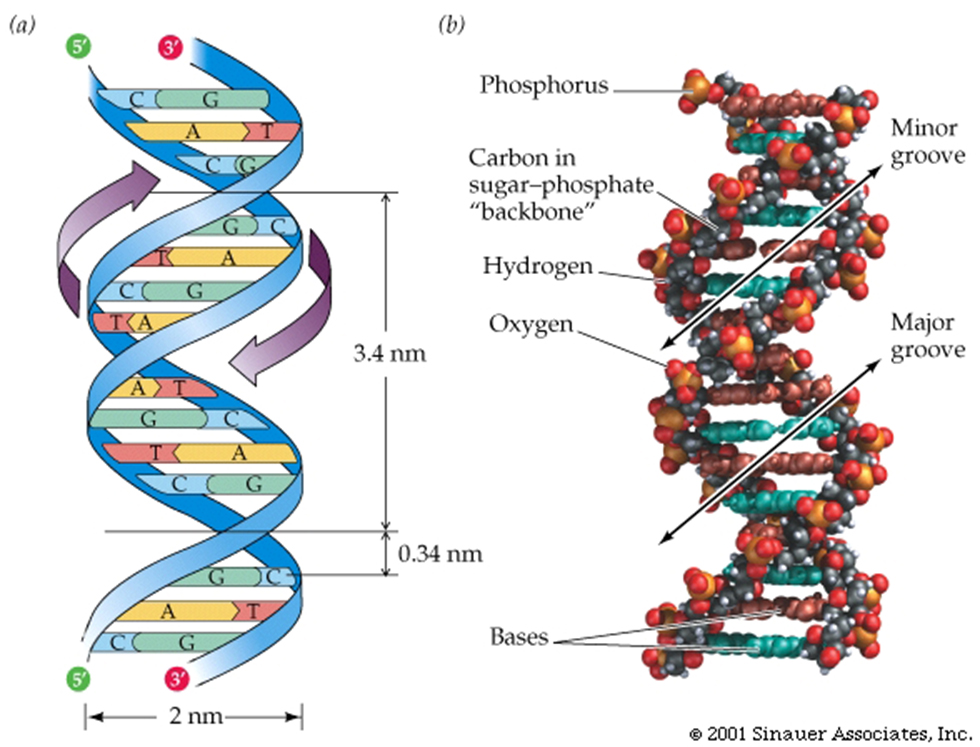

The DNA molecule is a double-stranded helix.The diameter of the DNA molecule is uniform, and easily interpreted by proteins that "patrol" the DNA to maintain it's integrity.
The twist in DNA is right-handed (-the same direction as the threads on most screws)...right handed double-helix, although the "threading" is not always the same.....B-form HelixThe two strands run in different directions (they are antiparallel).
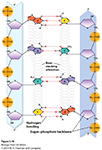
Hydrogen bonds -a type of attractive intermolecular force that exists between two partial electric charges of opposite polarity- between complementary bases hold the two strands together.
Consequently, each base pair has one purine and one pyrimidine, so the diameter of the double helix remains constant.
As the genetic material of the cell, DNA must perform four important functions:
It must be able to store all of an organism's genetic information.It must be susceptible to mutation.It must be precisely replicated in the cell division cycle.It must be expressible -giving rise to the phenotype.
In prokaryotes the replication complex makes new DNA at a rate in approximating 500 - 1,000 base pairs per second, with an error rate of fewer than one base in a billion.
In eukaryotes, the rate approximates 10 -fold less, but retains a similar replication error rate.
In both eukaryotes and prokaryotes, DNA is replicated by a huge enzyme complex, called DNA polymerase or DNA Replicase, as it catalyzes the reactions of DNA replication.
In eukaryotes, large chromosomes can have hundreds of origins of replication. Consequently, in eukaryotes, DNA replication occurs at many different sites simultaneously.
In both eukaryotes and prokaryotes, DNA is replicated by a large enzyme complex, called DNA polymerase or DNA Replicase, as it catalyzes the reactions of DNA replication.
Theoretically, DNA could serve as its own template in one of three different ways:

Semiconservative replication would use each parent strand as a template for a new strand. Each new DNA double helix would then have one parent strand and one new strand.
Conservative replication would build an entirely new double helix based on the template of the old double helix. The new strand would contain none of the original DNA.
Dispersive replication would use fragments of the original DNA molecule as templates for assembling two molecules. All the resulting strands would be mixtures of old and new material.
DNA polymerases are incredibly faithful in their replication porcess, with 1 error in 1x 109 bases replicated...., As a result, the process of DNA replication HAS to begin from an existing 3'OH group.... Consequently Replication of chromosomes by the Major replicase (DNA Polymerase) ALWAYS needs a "primer", which is a short strand of RNA complementary to the DNA template strand.
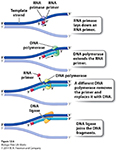
An enzyme called a primase makes the RNA primer strand.
The RNA primer is later degraded and replaced
with DNA, so the final DNA molecule has no RNA.
DNA helicase & topoisomerase: which denature and untangle the double helix.
Single-strand binding proteins: keeps the two strands separate.
RNA primase: makes a primer strand that serves as a starting point for replication.
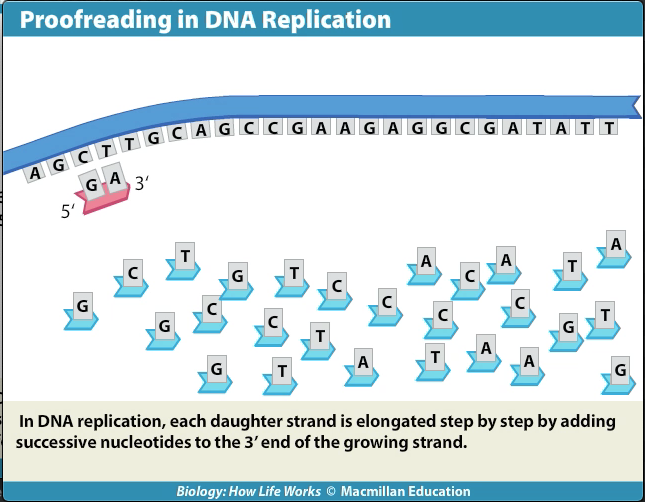
DNA polymerase: itself adds complementary nucleotides to the growing strand, proofreads the DNA, and repairs it.
DNA ligase: seals up breaks in the sugar–phosphate backbone.
So both strands cannot be synthesized in exactly the same way.
Because of its relatively opposite orientation, the lagging strand of DNA must grow in relatively small, discontinuous pieces, called Okazaki fragments. after their discoverer, the Japanese biochemist Reiji Okazaki.
Each Okazaki fragment requires an RNA primer to initiate strand synthesis, which is formed by an enzyme called RNA primase, which begins it synthesis of RNA some distance away from the previous Okazaki fragment.
Ultimately, the gaps in the lagging strand DNA are filled in by a repair DNA polymerase, which is also able to remove the RNA primers that are left on this strand.
Finally, DNA ligase catalyzes formation of the final phosphodiester linkage that joins the two Okazaki fragments.
Okazaki fragments range from around 100 to 200 nucleotides long in eukaryotes, and 1,000 to 2,000 nucleotides long in prokaryotes.
In E. coli, the replication complex makes new DNA at a rate in approximating 500 -1,000 base pairs per second, with an error rate of fewer than one base in a billion.
In Eukaryotes, the rate approximates 10 -fold less, but retains a similar replication error rate.
In both eukaryotes and prokaryotes, DNA is replicated by a huge enzyme complex, called DNA polymerase or DNA Replicase, as it catalyzes the reactions of DNA replication.

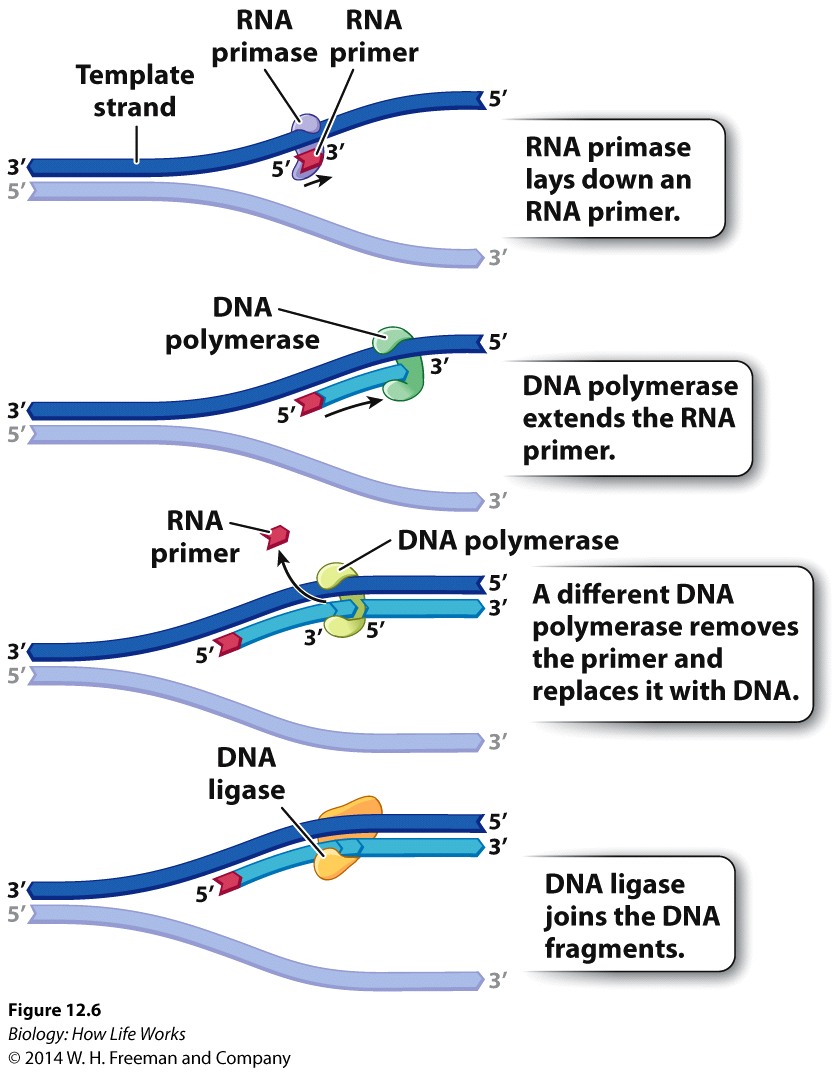

It is the RNA primase that always makes the primer strand, and as we have discussed, the resulting RNA primer is later degraded and replaced with DNA, so the final DNA molecule has no RNA.
As the genetic material, DNA must perform four important functions:
It must be able to store all of an organism's genetic information.It must be susceptible to mutation.It must be precisely replicated in the cell division cycle.It must be expressible as the phenotype.
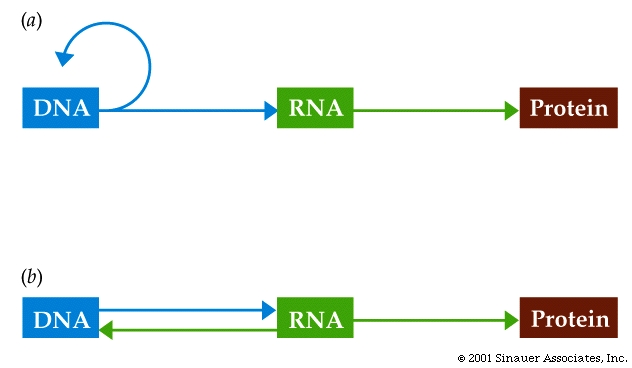
Central Dogma: DNA -> RNA -> Proteins!
There are many steps between genotype and phenotype; as genes cannot (by themselves) directly produce a phenotype.
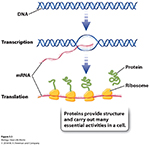

At the molecular level
- gene: "a sequence of DNA that gives rise to a functional gene product..." this product can (ultimately) be an RNA or a protein.
This is a little different from our first definition of a gene, way back in lecture 4, when we defined it as
- gene: A unit of heritable information -usually associated (at the molecular level) with a specific region located on the chromosome.
Given the central dogma, however, there are two steps which are commonly used to express a gene:
Transcription, which makes a single-stranded RNA copy of a segment of the DNA.
Followed by the process of Translation, which uses information encoded in a portion of the RNA to make a polypeptide.
In eukaryotes, these two steps are physically distinct... in prokaryotes -not so much.
How does RNA differ from DNA
RNA is usually single-stranded.The sugar in RNA is ribose, not deoxyribose.Wherever thymine is found in DNA, it is replaced by uracil in RNA.RNA can fold over and base-pair with itself.
RNA comes in various forms/sequences, commensurate with function.
rRNA (81% by weight in E. coli; cellular RNA), which acts as nucleic acid scaffold for the ribosomes, which are the enzymes that copy the mRNA message into a polypeptide chain.
tRNA (15% by weight in E. coli; 60 different possible species), which is the link between the code of the mRNA and the amino acids of the polypeptide. The tRNA molecules specify the correct amino acid.
mRNA (4% by weight in E. coli; transient 0.5-10 minute to 24hr. life span), which is the transient information that is copied from the DNA.
In addition to the traditional forms of RNA there is a 4th variety, known as Small interfering RNA (microRNA), which is a class of double-stranded RNA molecules of some 20-25 base pairs in length, which plays numerous roles in eukaryotes, but is most commonly understood to hybridize to specific regions of mRNA and "interfere" with its expression. They are often used as "antiviral" RNA's.
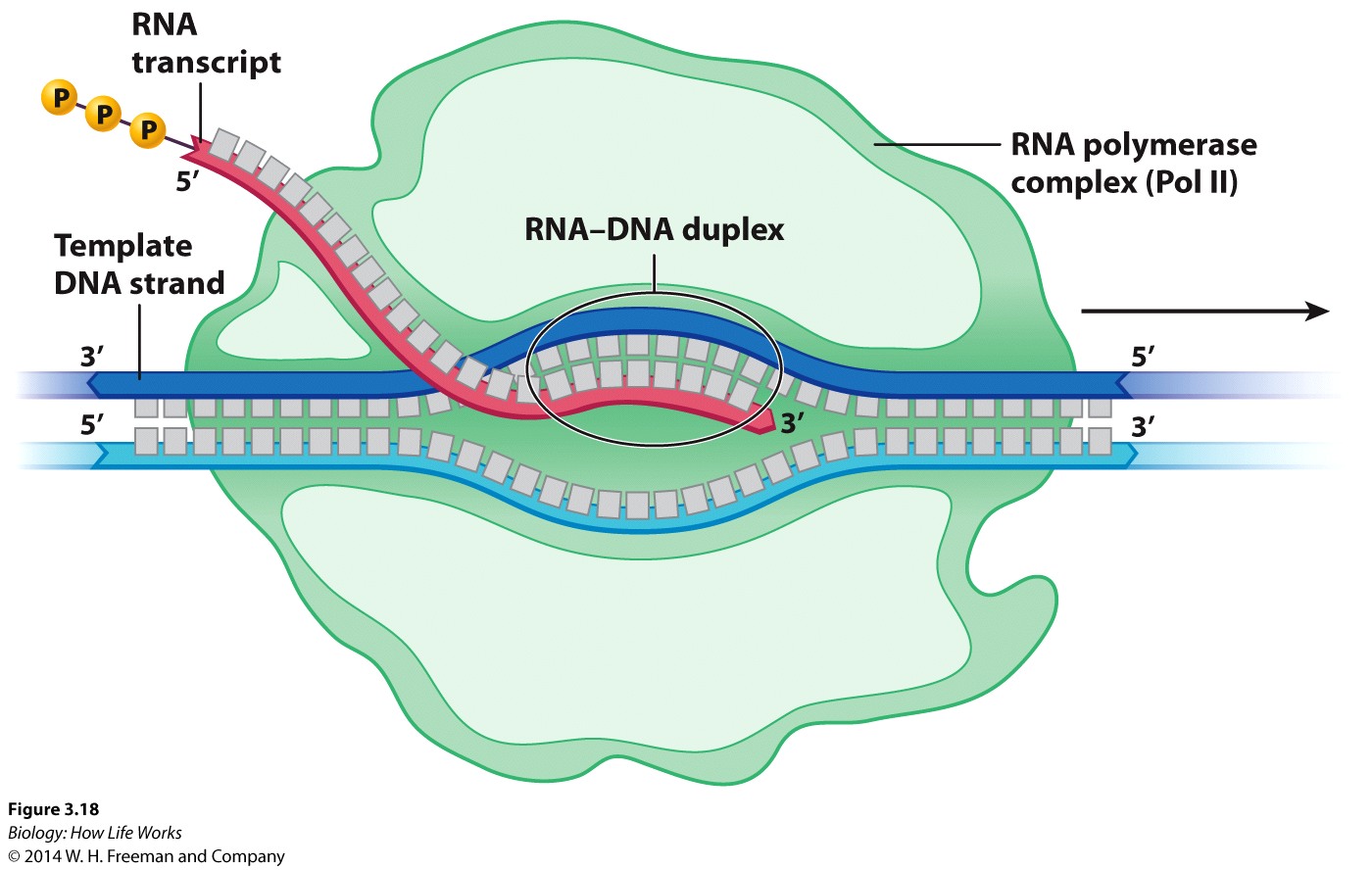
Synthesis of RNA:----- Transcription is a "DNA-Directed" process.
It is carried out by RNA polymerase, which is an enzyme that uses DNA as a template to make RNA.
Transcription of a gene begins at its promoter, which is a certain sequence of DNA that the RNA polymerase recognizes.

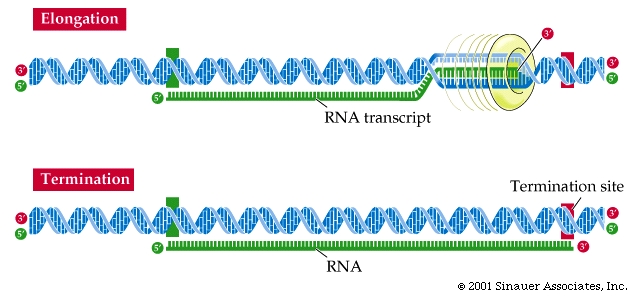
For any givent transcript, just one of the strands of a gene's DNA is used to make the RNA. This strand is called the template strand and is used for transcription. The other, untranscribed strand, is called the complementary strand.
For different genes in the same DNA molecule, however, the roles of these strands may be reversed.
Thus, the continuous double helix of DNA has many regions that are read by RNA polymerase, and needs to be partly unwound to serve as template.
As the RNA transcript forms, it peels away, allowing the already transcribed DNA to be rewound into the double helix.
Initiation of transcription requires a "promoter" and an RNA polymerase.
There is at least one "promoter" for each gene that is to be transcribed into mRNA.
The RNA polymerase binds to the promoter region when conditions allow.
The promoter sequence directs the RNA polymerase as to which of the double strands is the template and in what direction the RNA polymerase should move.
In effect, the promoters serve as "punctuation marks" for the transcriptional process.
As in replication of DNA, transcription of RNA is always synthesized in the 5' -to- 3' direction
Not all promoters are identical. Some bind RNA polymerase more effectively than others; which causes them to be transcribed more frequently........when conditions allow.
Prokaryotes have only ONE type of RNA polymerase which transcribes ALL THREE types of RNA, mRNA, tRNA, and rRNA.
Eukaryotes have THREE DIFFERENT RNA polymerases: RNA polymerase I, II, and III.
It is the RNA polymerase II that makes ALL the mRNA in eukaryotes.
In eukaryotic cells, other proteins must also bind to the DNA around the promoter to prepare a 'docking site' for the transcribing enzyme to bind. This uniquely eukaryotic feature provides a way to regulate the transcription of particular genes in a uniquely eukaryotic way.
RNA polymerase elongates the transcript.
After binding to the DNA template, RNA polymerase unwinds the DNA about 20 base pairs at a time and reads the template in the 3' -to- 5' direction (the opposite direction to RNA synthesis).
The RNA transcript is thus extended in an anti-parallel manner with respect to the DNA strand that acts as the "template" strand.
Much of the energy for synthesis comes from the removal and breakdown of the pyrophosphate group from each nucleotide added
Transcription errors for RNA polymerases are much higher (relative to DNA polymerases); making a mistake every 104 to 105 bases incorporated into the growing RNA strand.
Transcription terminates at particular base sequences in the DNA that specify termination.
RNA Transcription, and Gene expression in Eukaryotes is far more complex
https://www.youtube.com/watch?v=WsofH466lqk
Indeed, the whole Genetic Dogma is more complex in Eukaryotes......
The mRNA that gives rise to the protein product in prokaryotes is said to be co-linear with the DNA sequence from which it came.
This theme varies somewhat for eukaryotes.
1. 5' end of the mRNA is "capped" or modified to establish it as being the 5' end of a mRNA, eventually gives rise to the mature mRNA
2. Distinct lack of colinearity, and the existence of Exon and Intron sequences in the DNA and hnRNA.
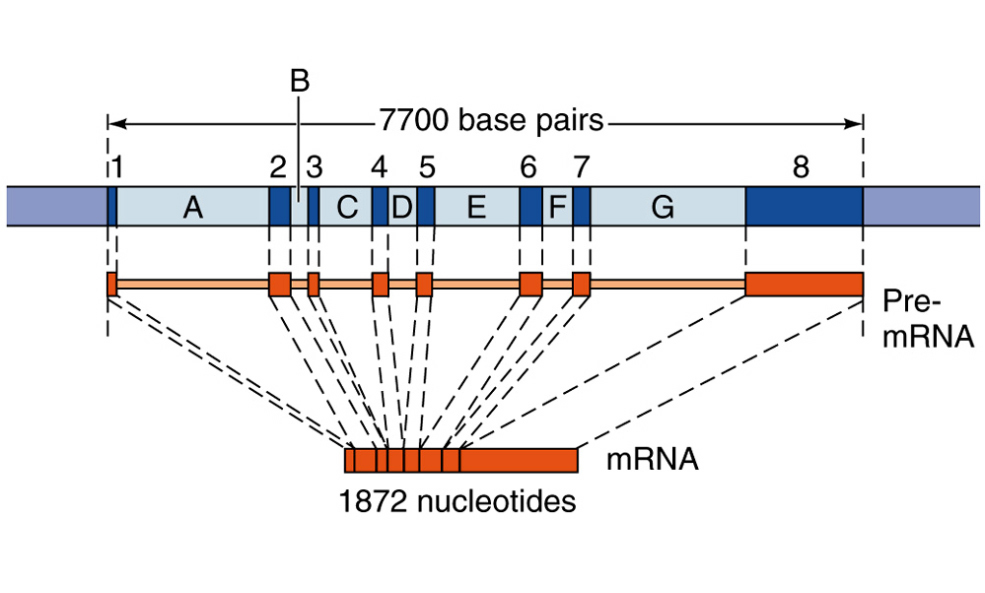
3. Very often mature mRNA sequences (or pre-RNA sequences) encode for a “3' untranslated region” (UTR) that (as the name suggests) is transcribed, but not translated.
This region includes the sequence 5'AATAAA3' (at the level of DNA), which, when transcribed in to RNA becomes is the signal for a PolyA tail (of approximately 200 to 300 nucleotides in higher mammals) to be attached to the maturing message
.thmb.jpg)
Consequently many of the major differences between Proks and Euks are defined by the separation of translation from the other aspects of the central dogma by the nuclear membrane.
In addition to being more complex in Eukaryotes than Prokaryotes, the Central Dogma is not entirely universal.
RNA viruses modify the central dogma, RNA is their information molecule during transmission.
Examples are the influenza virus and poliovirus.
HIV and certain tumor viruses have RNA as their infectious information molecule, but convert it to a DNA copy inside the host cell, then use it to make more RNA.
Translation of the RNA into polypeptides.
In prokaryotes, translation of the mRNA often begins before transcription is complete.
In eukaryotes, because the whole process is far more complicated and Replication, together with Transcription and maturation of the transcript is separated from Translation (in the cytoplasm or Edoplasmic Reticulum) by the nucelar membrane... translation can olny begin in Eukaryotes AFTER transcription and Maturation of the RNA is complete. Thereafter, however translation is very similar in both Eukaryotes except that in Eukaryotes the Ribosome complexes are a little bit larger than they are in Prokaryotes.
But let's start with the basics.
The Genetic Code
DNA codes for RNA through the transcriptional process.
mRNA is a single stranded nucelic acid that is converted into a polypeptide chain through a "3 for one" genetic code. The mRNA is "read" in three-base, contiguous segments called codons (equivalent to "words").
The number of different codons possible is 64, because each position in the codon can be occupied by one of four different bases.



Four possibilities for the first base, times four for the second, times four for the third yields 64 possibilities.
In the genetic code, there are no commas or punctuation marks,it is therefore very important to know where to BEGIN and where to END the process. For this reason there are always well defined START and STOP "codons".
L / ETT / HEC / ATG / ETT / HER / ATO / FFT / HEB / AT
LE / TTH / ECA / TGE / TTH / ERA/ TOF / FTH / EBA / T
LET / THE / CAT / GET / THE / RAT / OFF / THE / BAT
The 64 possible codons (or "words") code for only 20 amino acids as well as the start and stop signals found in ALL mRNA molecules.
AUG, which codes for the amino acid methionine, is called the start codon, which initiates the translational process.
Three of the possible codons are stop< codons (UAA, UAG, and UGA), which direct the ribosomes to STOP reading the mRNA; that is, they end translation.
The
genetic code is redundant but
not ambiguous. This means
that many amino acids have more than one
codon, but only one amino acid
is specified for any one codon.
The genetic code is nearly Universal, applying to all species on our planet. (which would argue strongly for an evolution. Why?? )
Minor variations are found, however, within mitochondria and chloroplasts; but all other exceptions are few and far between.
Translation occurs at ribosomes, which are molecular protein synthesizing machines that hold mRNA and tRNA in place.